Tips to speed up Python¶
Contents¶
Hang on, don’t optimise too early¶
Trade-offs e.g. complexity, speed, memory, disk, readability, time, effort, etc.
Check that code is correct (tested, documented).
Is optimisation needed?
If yes, optimise code and data.
If more needed, parallelise.
Plot idea from Dask-ML.
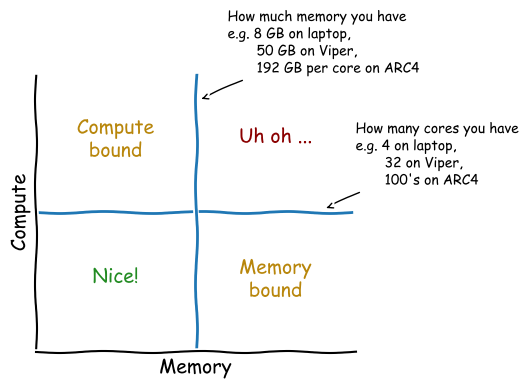
How fast is it?¶
Profiling (i.e. find the bottlenecks)
-
IPython magic (Jupyter Lab)
Line:
%timeitCell:
%%timeitIf
pip install line_profiler:First load module:
%load_ext line_profilerScripts:
%prunLine-by-line:
%lprun@profiledecorator around the function
-
If
pip install memory_profiler:First load module:
%load_ext memory_profilerLine:
%memitCell:
%%memitLine-by-line:
%mprun
-
How fast could it go?¶
Time-space complexity
Big O notation where O is the order of operations, O(…).
Ignores constants and takes the largest order, so O(2n2 + 3n) would be O(n2).
Important for large number of elements, N.
Typical case.
Constant time means per machine operation.
Plot idea from Big O Cheat Sheet
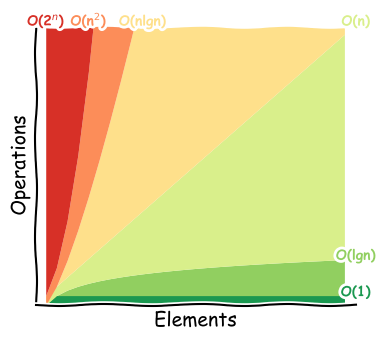
Potential improvements¶
Append to lists, rather than concatenating¶
Lists are allocated twice the memory required, so appending fills this up in O(1) (long-term average), while concatenating creates a new list each time in O(n).
%%timeit
my_list = []
for num in range(1_000):
my_list += [num] # time O(n)
46.5 µs ± 76.3 ns per loop (mean ± std. dev. of 7 runs, 10000 loops each)
%%timeit
my_list = []
for num in range(1_000):
my_list.append(num) # time O(1)
35.6 µs ± 130 ns per loop (mean ± std. dev. of 7 runs, 10000 loops each)
Move loop-invariants outside loops¶
%%timeit
for num in range(1_000_000):
constant = 500_000
bigger_num = max(num, constant)
124 ms ± 462 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
%%timeit
constant = 500_000
for num in range(1_000_000):
bigger_num = max(num, constant)
115 ms ± 196 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Use built-in functions¶
Optimised in C (statically typed and compiled).
nums = [num for num in range(1_000_000)]
%%timeit
count = 0
for num in nums: # time O(n)
count += 1
23.8 ms ± 920 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
%timeit len(nums) # time O(1)
42.7 ns ± 2.1 ns per loop (mean ± std. dev. of 7 runs, 10000000 loops each)
Use optimal data types¶
Common and additional data structures.
e.g. dictionaries are fast to search, O(1).
Example from Luciano Ramalho, Fluent Python, Clear, Concise, and Effective Programming, 2015. O’Reilly Media, Inc.
haystack_list = np.random.uniform(low=0, high=100, size=(1_000_000))
haystack_dict = {key: value for key, value in enumerate(haystack_list)}
needles = [0.1, 50.1, 99.1]
%%timeit
needles_found = 0
for needle in needles:
if needle in haystack_list: # time O(n) within list
needles_found += 1
602 µs ± 13.5 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
%%timeit
needles_found = 0
for needle in needles:
if needle in haystack_dict: # time O(1) within dict
needles_found += 1
138 ns ± 0.213 ns per loop (mean ± std. dev. of 7 runs, 10000000 loops each)
Many more examples e.g.:
Generators save memory by yielding only the next iteration.
Memory usage for floats/integers of 16 bit < 32 bit < 64 bit.
For NetCDFs, using
engine='h5netcdf'withxarraycan be faster, over than the defaultengine='netcdf4'.Compression: If arrays are mostly 0, then can save memory using sparse arrays.
Chunking: If need all data, then can load/process in chunks to reduce amount in memory: Zarr for arrays, Pandas.
Indexing: If need a subset of the data, then can index (multi-index) to reduce memory and increase speed for queries: Pandas, SQLite.
Reduce repeated calculations with caching¶
e.g. Fibonacci sequence (each number is the sum of the two preceding ones starting from 0 and 1 e.g. 0, 1, 1, 2, 3, 5, 8, 13, 21, 34).
def fibonacci(n): # time O(2^n) as 2 calls to the function n times (a balanced tree of repeated calls)
if n == 0 or n == 1:
return 0
elif n == 2:
return 1
else:
return fibonacci(n - 1) + fibonacci(n - 2)
%timeit fibonacci(20)
1.04 ms ± 6 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
def fibonacci_with_caching(n, cache={0: 0, 1: 0, 2: 1}): # time O(n) as 1 call per n
if n in cache:
return cache[n]
else:
cache[n] = fibonacci_with_caching(n - 1, cache) + fibonacci_with_caching(n - 2, cache)
return cache[n]
%timeit fibonacci_with_caching(20, cache={0: 0, 1: 0, 2: 1})
4.23 µs ± 60.2 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)
Use vectorisation instead of loops¶
Loops are slow in Python (CPython, default interpreter).
Because loops type−check and dispatch functions per cycle.
Vectors can work on many parts of the problem at once.
NumPy ufuncs (universal functions).
Optimised in C (statically typed and compiled).
nums = np.arange(1_000_000)
%%timeit
for num in nums:
num *= 2
274 ms ± 1.28 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
%%timeit
double_nums = np.multiply(nums, 2)
617 µs ± 41 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
Broadcasting (ufuncs over different shaped arrays, NumPy, xarray).

nums_col = np.array([0, 10, 20, 30]).reshape(4, 1)
nums_row = np.array([0, 1, 2])
nums_col + nums_row
array([[ 0, 1, 2],
[10, 11, 12],
[20, 21, 22],
[30, 31, 32]])
import xarray as xr
nums_col = xr.DataArray([0, 10, 20, 30], [('col', [0, 10, 20, 30])])
nums_row = xr.DataArray([0, 1, 2], [('row', [0, 1, 2])])
nums_col + nums_row
<xarray.DataArray (col: 4, row: 3)>
array([[ 0, 1, 2],
[10, 11, 12],
[20, 21, 22],
[30, 31, 32]])
Coordinates:
* col (col) int64 0 10 20 30
* row (row) int64 0 1 2- col: 4
- row: 3
- 0 1 2 10 11 12 20 21 22 30 31 32
array([[ 0, 1, 2], [10, 11, 12], [20, 21, 22], [30, 31, 32]]) - col(col)int640 10 20 30
array([ 0, 10, 20, 30])
- row(row)int640 1 2
array([0, 1, 2])
Algorithm improvements¶
The instructions to solve the problem.
Free MIT course on ‘Introduction to algorithms’, video lectures.
Many existing libraries are already optimised (computationally and algorithmically).
Minimal examples of data structures and algorithms in Python.
e.g. Find multiple strings in a text.
Aho-Corasick algorithm, 25x faster than using regex naively.
Compilers¶
-
Ahead-Of-Time (AOT) compiler.
Statically compiled C extensions.
General purpose interpreter.
Can work on a variety of problems.
Dynamically typed.
Types can change e.g.
x = 5, then laterx = 'gary'.
-
Just−In−Time (JIT) compiler (written in Python).
Enables optimisations at run time, especially for numerical tasks with repitition and loops.
Replaces CPython.
Faster, though overheads for start-up and memory.
from numba import njit
nums = np.arange(1_000_000)
def super_function(nums):
trace = 0.0
for num in nums: # loop
trace += np.cos(num) # numpy
return nums + trace # broadcasting
%timeit super_function(nums)
1.68 s ± 8.53 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
@njit # numba decorator
def super_function(nums):
trace = 0.0
for num in nums: # loop
trace += np.cos(num) # numpy
return nums + trace # broadcasting
%timeit super_function(nums)
15 ms ± 46 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
-
Compiles to statically typed C/C++.
Use for any amount of code.
Use with the default CPython.
Examples not using IPython, NumPy, Pandas (example below).
import pandas as pd
df = pd.DataFrame({
"a": np.random.randn(1000),
"b": np.random.randn(1000),
"N": np.random.randint(100, 1000, (1000)),
"x": "x",
})
df.head()
| a | b | N | x | |
|---|---|---|---|---|
| 0 | -1.860311 | -0.551809 | 288 | x |
| 1 | 0.782267 | 1.564407 | 653 | x |
| 2 | -1.243118 | -0.414263 | 472 | x |
| 3 | 0.107798 | -1.261014 | 312 | x |
| 4 | -0.524740 | 0.479698 | 958 | x |
def f(x):
return x * (x - 1)
def integrate_f(a, b, N):
s = 0
dx = (b - a) / N
for i in range(N):
s += f(a + i * dx)
return s * dx
%timeit df.apply(lambda x: integrate_f(x["a"], x["b"], x["N"]), axis=1)
73 ms ± 794 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
%load_ext Cython
%%cython # only change
def f(x):
return x * (x - 1)
def integrate_f(a, b, N):
s = 0
dx = (b - a) / N
for i in range(N):
s += f(a + i * dx)
return s * dx
%timeit df.apply(lambda x: integrate_f(x["a"], x["b"], x["N"]), axis=1)
51.7 ms ± 729 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
%%cython
cdef double f(double x) except? -2: # adding types
return x * (x - 1)
cpdef double integrate_f(double a, double b, int N): # adding types
cdef int i # adding types
cdef double s, dx # adding types
s = 0
dx = (b - a) / N
for i in range(N):
s += f(a + i * dx)
return s * dx
%timeit df.apply(lambda x: integrate_f(x["a"], x["b"], x["N"]), axis=1)
8.96 ms ± 106 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Lazy loading and execution¶
Lazily loads metadata only, rather than eagerly loading data into memory.
Creates task graph of scheduled work awaiting execution (
.compute()).
xr.tutorial.open_dataset('air_temperature')
<xarray.Dataset>
Dimensions: (lat: 25, lon: 53, time: 2920)
Coordinates:
* lat (lat) float32 75.0 72.5 70.0 67.5 65.0 ... 25.0 22.5 20.0 17.5 15.0
* lon (lon) float32 200.0 202.5 205.0 207.5 ... 322.5 325.0 327.5 330.0
* time (time) datetime64[ns] 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 ...
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...- lat: 25
- lon: 53
- time: 2920
- lat(lat)float3275.0 72.5 70.0 ... 20.0 17.5 15.0
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
array([75. , 72.5, 70. , 67.5, 65. , 62.5, 60. , 57.5, 55. , 52.5, 50. , 47.5, 45. , 42.5, 40. , 37.5, 35. , 32.5, 30. , 27.5, 25. , 22.5, 20. , 17.5, 15. ], dtype=float32) - lon(lon)float32200.0 202.5 205.0 ... 327.5 330.0
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
array([200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. ], dtype=float32) - time(time)datetime64[ns]2013-01-01 ... 2014-12-31T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]')
- air(time, lat, lon)float32...
- long_name :
- 4xDaily Air temperature at sigma level 995
- units :
- degK
- precision :
- 2
- GRIB_id :
- 11
- GRIB_name :
- TMP
- var_desc :
- Air temperature
- dataset :
- NMC Reanalysis
- level_desc :
- Surface
- statistic :
- Individual Obs
- parent_stat :
- Other
- actual_range :
- [185.16 322.1 ]
[3869000 values with dtype=float32]
- Conventions :
- COARDS
- title :
- 4x daily NMC reanalysis (1948)
- description :
- Data is from NMC initialized reanalysis (4x/day). These are the 0.9950 sigma level values.
- platform :
- Model
- references :
- http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanalysis.html
Parallelisation¶
Parallelisation divides a large problem into many smaller ones and solves them simultaneously.
Divides up the time/space complexity across workers.
Tasks centrally managed by a scheduler.
Multi-processing (cores) - useful for compute-bound problems.
Multi-threading (parts of processes), useful for memory-bound problems.
If need to share memory across chunks:
Use shared memory (commonly OpenMP, Open Multi-Processing).
-pe smp npon ARC4
Otherwise:
Use message passing interface, MPI (commonly OpenMPI).
-pe ib npon ARC4
Single machine¶
See the excellent video from Dask creator, Matthew Rocklin, below.
-
Great features.
Helpful documentation.
Familiar API.
Under the hood for many libraries e.g. xarray, iris, scikit-learn.
from dask.distributed import Client
client = Client()
client
ds = xr.open_dataset(
'/nfs/a68/shared/earlacoa/wrfout_2015_PM_2_DRY_0.25deg.nc',
chunks={'time': 'auto'} # dask chunks
)
ds.nbytes * (2 ** -30)
%time ds_mean = ds.mean()
%time ds_mean.compute()
ds.close()
client.close()
Multi-threading¶
See the excellent video from Dask creator, Matthew Rocklin, below.
e.g. dask.array (NumPy).
from dask.distributed import Client
client = Client(
processes=False,
threads_per_worker=4,
n_workers=1
)
client
import dask.array as da
my_array = da.random.random(
(50_000, 50_000),
chunks=(5_000, 5_000) # dask chunks
)
result = my_array + my_array.T
result
result.compute()
client.close()
Multi-processing¶
See the excellent video from Dask creator, Matthew Rocklin, below.
e.g. dask.dataframe (Pandas).
from dask.distributed import Client
client = Client()
client
import dask
import dask.dataframe as dd
df = dask.datasets.timeseries()
df
type(df)
result = df.groupby('name').x.std()
result
result.visualize()
result_computed = result.compute()
type(result_computed)
client.close()
Interactive on HPC¶
See the excellent video from Dask creator, Matthew Rocklin, below.
Create or edit the
~/.config/dask/jobqueue.yamlfile with that in this directory.Also, can check the
~/.config/dask/distributed.yamlfile with that in this directory.
e.g. dask.bag
Iterate over a bag of independent objects (embarrassingly parallel).
# in a terminal
# log onto arc4
ssh ${USER}@arc4.leeds.ac.uk
# start an interactive session on a compute node on arc4
qlogin -l h_rt=04:00:00 -l h_vmem=12G
# activate your python environment
conda activate my_python_environment
# echo back the ssh command to connect to this compute node
echo "ssh -N -L 2222:`hostname`:2222 -L 2727:`hostname`:2727 ${USER}@arc4.leeds.ac.uk"
# launch a jupyter lab session on this compute node
jupyter lab --no-browser --ip=`hostname` --port=2222
# in a local terminal
# ssh into the compute node
ssh -N -L 2222:`hostname`:2222 -L 2727:`hostname`:2727 ${USER}@arc4.leeds.ac.uk
# open up a local browser (e.g. chrome)
# go to the jupyter lab session by pasting into the url bar
localhost:2222
# can also load the dask dashboard in the browser at localhost:2727
# now the jupyter code
from dask_jobqueue import SGECluster
from dask.distributed import Client
cluster = Client(
walltime='01:00:00',
memory='4 G',
resource_spec='h_vmem=4G',
scheduler_options={
'dashboard_address': ':2727',
},
)
client = Client(cluster)
cluster.scale(jobs=20)
#cluster.adapt(minimum=0, maximum=20)
client
import numpy as np
import dask.bag as db
nums = np.random.randint(low=0, high=100, size=(5_000_000))
nums
def weird_function(nums):
return chr(nums)
bag = db.from_sequence(nums)
bag = bag.map(weird_function)
bag.visualize()
result = bag.compute()
client.close()
cluster.close()
HPC¶
Non-interactive.
Create/edit the
dask_on_hpc.pyfile.Submit to the queue using
qsub dask_on_hpc.bash.
Profile parallel code¶
Recommendations¶
Check code is correct and optimisation is needed.
Profile to find bottlenecks.
Jupyter Lab:
%%timeitand%%memit.Line-by-line:
line_profilerandmemory_profiler.
Optimise code and data.
Parallelise.
Further information¶
Helpful resources
Why is Python slow?, Anthony Shaw, PyCon 2020. CPython Internals.
Luciano Ramalho, Fluent Python, Clear, Concise, and Effective Programming, 2015. O’Reilly Media, Inc.
Jake VanderPlas, Python Data Science Handbook, 2016. O’Reilly Media, Inc.
Pangeo, talk - Python libraries that work well together and build on each other, especially for big data geosciences (e.g. NumPy, Pandas, xarray, Dask, Numba, Jupyter).
Concurrency can also run different tasks together, but work is not done at the same time.
Asynchronous (multi-threading), useful for massive scaling, threads controlled explicitly.
MIT course on ‘Introduction to algorithms’, video lectures.
PythonSpeed.com, Itamar Turner-Trauring
Writing faster Python, Sebastian Witowski, Euro Python 2016
Other things that may help save time in the long run:
